By Michel Danino
(Published in Puratattva, Bulletin of the Indian Archaeological Society, New Delhi, No. 36, 2005-06, pp. 146-154.)
Background
Along with the birth of anthropology, the nineteenth century saw the development of semi-scientific to wholly unscientific disciplines, such as anthropometry, craniometry or phrenology. Unquestioningly accepting the prevalent concept of race, some scientists constructed facial and nasal indexes or claimed to measure the skull’s volume for every race, of course with the result that the white race’s cranium was the most capacious and its owner, therefore, the most intelligent; others went further, insisting that amidst the white race, only the Germans were the “pure” descendants of the “Aryan race” which was destined the rule the earth.
In India, from 1891 onward, Herbert H. Risley, an official with the colonial government, set about defining in all seriousness 2,378 castes belonging to 43 “races,” all of it on the basis of a “nasal index.” The main racial groups were Indo-Aryan, Turko- Iranian, Scytho-Dravidian, Aryo-Dravidian, Mongoloid and Mongolo-Dravidian.
Unfortunately, this imaginative but wholly unscientific work weighed heavily on the first developments of Indian anthropology; in the 1930s, for instance, B. S. Guha studied skeletons from Mohenjo-daro and submitted a detailed report on the proto- Australoid, Mediterranean, Mongoloid and Alpine races peopling the city, all of them “non-Aryan” of course. Long lists of such fictitious races filled academic publications, and continue to be found in Indian textbooks today.
In the wake of World War II, the concept of race collapsed in the West. Rather late in the day, anthropologists realized that race cannot be scientifically defined, much less measured, thus setting at naught a whole century of scholarly divagations on “superior” and “inferior” races. Following in the footsteps of pioneers like Franz Boas,1 leading scientists, such as Ashley Montagu,2 now argued strongly against the “fallacy of race.” It is only with the emergence of more reliable techniques in biological anthropology that anthropometry got a fresh chance; it concentrated not on trying to categorize noses or spot “races,” but on tracing the evolution of a population, on signs of continuity or disruption, and on possible kinships between neighbouring populations.
In the Indian context, we are now familiar with the work of U.S anthropologists Kenneth Kennedy, John Lukacs and Brian Hemphill.3 Their chief conclusion, as far as the Aryan debate is concerned, is that there is no trace of “demographic disruption” in the North-West of the subcontinent between 4500 and 800 BCE; this negates the possibility of any massive intrusion, by so-called Indo-Aryans or other populations, during that period.
Die-hard proponents of such an invasion / migration have therefore been compelled to downscale it to a “trickle-in” infiltration,4 limited enough to have left no physical trace, although they are at pains to explain how a “trickle” was able to radically alter India’s linguistic and cultural landscape when much more massive invasions of the historical period failed to do so.5 Other proponents still insist that “the Indo-Aryan immigrants seem to have been numerous and strong enough to continue and disseminate much of their culture,”6 but do not explain how the “immigrants” failed to leave any trace in the anthropological record.
A powerful new tool
In the 1980s, another powerful tool of inquiry came on the scene: genetics, with its growing ability to read the history contained in a human body’s three billion bits of information. In particular, techniques used in the identification of genetic markers have been fast improving, leading to a wide array of applications, from therapeutics to crime detection to genealogy. Let us first summarize the basic definitions relevant to our field.
In trying to reconstruct ancestry, biologists use two types of DNA, the complex molecule that carries genetic information. The first, Y-DNA, is contained in the Y- chromosome, one of the two sex chromosomes; it is found in the cell’s nucleus and is transmitted from father to son. The second, mtDNA or mitochondrial DNA, is found in mitochondria, kinds of power generators found in a cell, but outside its nucleus; this mtDNA is independent of the Y-DNA, simpler in structure, and transmitted by the mother alone. For various reasons, all this genetic material undergoes slight alterations or “mutations” in the course of time; those mutations then become characteristic of the line of descendants: if, for instance, the mtDNAs of two humans, however distant geographically, exhibit the same mutation, they necessarily share a common ancestor in the maternal line.
Much of the difficulty lies in organizing those mutations, or genetic markers, in consistent categories called “haplotypes” (from a Greek word meaning “single”), which constitute an individual’s genetic fingerprint. Similar haplotypes are then brought together in “haplogroups,” each of which genetically identifies a particular ethnic group. Such genetic markers can then be used to establish a “genetic distance” between two populations.
Identifying and making sense of the right genetic markers is not the only difficulty; dating their mutations remains a major challenge: on average, a marker of Y- DNA may undergo one mutation every 500 generations, but sudden changes caused by special circumstances can never be ruled out. Genetics, therefore, needs the inputs from palaeontology and archaeology, among other disciplines, to confirm its historical conclusions.
India’s case
Since the 1990s, there have been numerous genetic studies of Indian populations, often reaching apparently divergent conclusions. There are three reasons for this: (1) the Indian region happens to be one of the most diverse and complex in the world, which makes it difficult to interpret the data; (2) early studies relied on too limited samples, of the order of a few dozens, when hundreds or ideally thousands of samples are required for some statistical reliability; (3) some of the early studies fell into the old trap of trying to equate linguistic groups with distinct ethnic entities — a relic of the nineteenth-century erroneous identification between language and race; as a result, a genetic connection between North Indians and Central Asians was automatically taken to confirm an Aryan invasion in the second millennium BCE, disregarding a number of alternative explanations.7
More recent studies, using larger samples and much refined methods of analysis, both at the conceptual level and in the laboratory, have reached very different conclusions (interestingly, some of their authors had earlier gone along with the old Aryan paradigm8). We will summarize here the chief results of nine studies from various Western and Indian Universities, most of them conducted by international teams of biologists, and more than half of them in the last three years; since their papers are complex and technical, what follows is, necessarily, highly simplified and represents only a small part of their content.
The first such study dates back to 1999 and was conducted by the Estonian biologist Toomas Kivisild, a pioneer in the field, with fourteen co-authors from various nationalities (including M. J. Bamshad).9 It relied on 550 samples of mtDNA and identified a haplogroup called “U” as indicating a deep connection between Indian and Western-Eurasian populations. However, the authors opted for a very remote separation of the two branches, rather than a recent population movement towards India; in fact, “the subcontinent served as a pathway for eastward migration of modern humans” from Africa, some 40,000 years ago:
“We found an extensive deep late Pleistocene genetic link between contemporary Europeans and Indians, provided by the mtDNA haplogroup U, which encompasses roughly a fifth of mtDNA lineages of both populations. Our estimate for this split [between Europeans and Indians] is close to the suggested time for the peopling of Asia and the first expansion of anatomically modern humans in Eurasia and likely pre-dates their spread to Europe.”
In other words, the timescale posited by the Aryan invasion / migration framework is inadequate, and the genetic affinity between the Indian subcontinent and Europe “should not be interpreted in terms of a recent admixture of western Caucasoids10 with Indians caused by a putative Indo-Aryan invasion 3,000–4,000 years BP.”
The second study was published just a month later. Authored by U.S. biological anthropologist Todd R. Disotell,11 it dealt with the first migration of modern man from Africa towards Asia, and found that migrations into India “did occur, but rarely from western Eurasian populations.” Disotell made observations very similar to those of the preceding paper:
“The supposed Aryan invasion of India 3,000–4,000 years before present therefore did not make a major splash in the Indian gene pool. This is especially counter-indicated by the presence of equal, though very low, frequencies of the western Eurasian mtDNA types in both southern and northern India. Thus, the ‘caucasoid’ features of south Asians may best be considered ‘pre-caucasoid’ — that is, part of a diverse north or north-east African gene pool that yielded separate origins for western Eurasian and southern Asian populations over 50,000 years ago.”
Here again, the Eurasian connection is therefore traced to the original migration out of Africa. On the genetic level, “the supposed Aryan invasion of India 3000-4000 years ago was much less significant than is generally believed.”
A year later, thirteen Indian scientists led by Susanta Roychoudhury studied 644 samples of mtDNA from some ten Indian ethnic groups, especially from the East and South.12 They found “a fundamental unity of mtDNA lineages in India, in spite of the extensive cultural and linguistic diversity,” pointing to “a relatively small founding group of females in India.” Significantly, “most of the mtDNA diversity observed in Indian populations is between individuals within populations; there is no significant structuring of haplotype diversity by socio-religious affiliation, geographical location of habitat or linguistic affiliation.” That is a crucial observation, which later studies will endorse: on the maternal side at least, there is no such thing as a “Hindu” or “Muslim” genetic identity, nor even a high- or low-caste one, a North- or South-Indian one — hence the expressive title of the study: “Fundamental genomic unity of ethnic India is revealed by analysis of mitochondrial DNA.”
The authors also noted that haplogroup “U,” already noted by Kivisild et al. as being common to North Indian and “Caucasoid” populations, was found in tribes of eastern India such as the Lodhas and Santals, which would not be the case if it had been introduced through Indo-Aryans. Such is also the case of the haplogroup “M,” another marker frequently mentioned in the early literature as evidence of the invasion: in reality, “we have now shown that indeed haplogroup M occurs with a high frequency, averaging about 60%, across most Indian population groups, irrespective of geographical location of habitat. We have also shown that the tribal populations have higher frequencies of haplogroup M than caste populations.”
Also in 2000, twenty authors headed by Kivisild contributed a chapter to a book on the “archaeogenetics” of Europe.13 They first stressed the importance of the mtDNA haplogroup “M” common to India (with a frequency of 60%), Central and Eastern Asia (40% on average), and even to American Indians; however, this frequency drops to 0.6% in Europe, which is “inconsistent with the ‘general Caucasoidness’ of Indians.”
This shows, once again, that “the Indian maternal gene pool has come largely through an autochthonous history since the Late Pleistocene.” The authors then studied the “U” haplogroup, finding its frequency to be 13% in India, almost 14% in North-West Africa, and 24% from Europe to Anatolia; but, in their opinion, “Indian and western Eurasian haplogroup U varieties differ profoundly; the split has occurred about as early as the split between the Indian and eastern Asian haplogroup M varieties. The data show that both M and U exhibited an expansion phase some 50,000 years ago, which should have happened after the corresponding splits.” In other words, there is a genetic connection between India and Europe, but a far more ancient one than was thought.
Another important point is that looking at mtDNA as a whole, “even the high castes share more than 80 per cent of their maternal lineages with the lower castes and tribals”; this obviously runs counter to the invasionist thesis. Taking all aspects into consideration, the authors conclude: “We believe that there are now enough reasons not only to question a ‘recent Indo-Aryan invasion’ into India some 4000 BP, but alternatively to consider India as a part of the common gene pool ancestral to the diversity of human maternal lineages in Europe.” Mark the word “ancestral.”
After a gap of three years, Kivisild directed two fresh studies. The first, with nine
colleagues, dealt with the origin of languages and agriculture in India.14 Those biologists stressed India’s genetic complexity and antiquity, since “present-day Indians [possess] at least 90 per cent of what we think of as autochthonous Upper Palaeolithic maternal lineages.” They also observed that “the Indian mtDNA tree in general [is] not subdivided according to linguistic (Indo-European, Dravidian) or caste affiliations,” which again demonstrates the old error of conflating language and race or ethnic group.
Then, in a new development, they punched holes in the methodology followed by studies basing themselves on the Y-DNA (the paternal line) to establish the Aryan invasion, and point out that if one were to extend their logic to populations of Eastern and Southern India, one would be led to an exactly opposite result: “the straightforward suggestion would be that both Neolithic (agriculture) and Indo-European languages arose in India and from there, spread to Europe.” The authors do not defend this thesis, but simply guard against “misleading interpretations” based on limited samples and faulty methodology.
The second study of 2003, a particularly detailed one dealing with the genetic heritage of India’s earliest settlers, had seventeen co-authors with Kivisild (including L. Cavalli-Sforza and P. A. Underhill), and relied on nearly a thousand samples from the subcontinent, including two Dravidian-speaking tribes from Andhra Pradesh.15 Among other important findings, it stressed that the Y-DNA haplogroup “M17,” regarded till recently as a marker of the Aryan invasion, and indeed frequent in Central Asia, is equally found in the two tribes under consideration, which is inconsistent with the invasionist framework. Moreover, one of the two tribes, the Chenchus, is genetically close to several castes, so that there is a “lack of clear distinction between Indian castes and tribes,” a fact that can hardly be overemphasized.
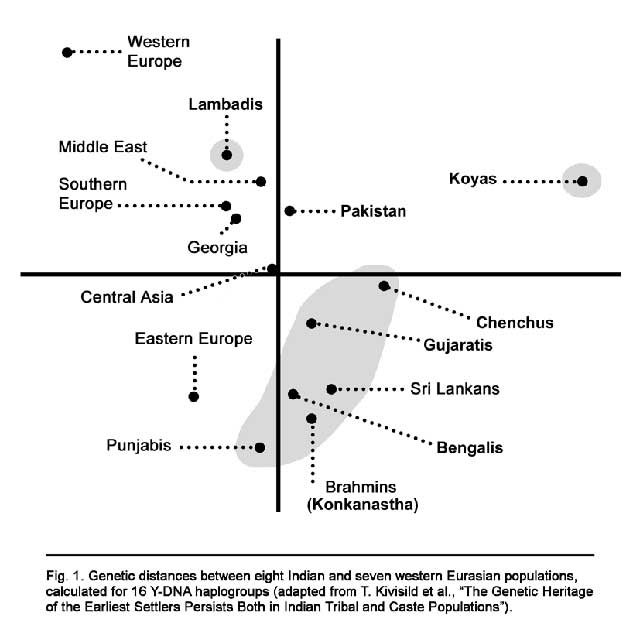
![genetic map]()
This also emerges from a diagram of genetic distances between eight Indian and seven Eurasian populations, distances calculate on the basis of 16 Y-DNA haplogroups (Fig. 1). The diagram challenges many common assumptions: as just mentioned, five castes are grouped with the Chenchus; another tribe, the Lambadis (probably of Rajasthani origin), is stuck between Western Europe and the Middle East; Bengalis of various castes are close to Mumbai Brahmins, and Punjabis (whom one would have thought to be closest to the mythical “Aryans”) are as far away as possible from Central Asia! It is clear that no simple framework can account for such complexity, least of all the Aryan invasion / migration framework.The next year, Mait Metspalu and fifteen co-authors analyzed 796 Indian (including both tribal and caste populations from different parts of India) and 436 Iranian mtDNAs.16 Of relevance here is the following observation, which once again highlights the pitfalls of any facile ethnic-linguistic equation:
“Language families present today in India, such as Indo-European, Dravidic and Austro-Asiatic, are all much younger than the majority of indigenous mtDNA lineages found among their present-day speakers at high frequencies. It would make it highly speculative to infer, from the extant mtDNA pools of their speakers, whether one of the listed above linguistically defined group in India should be considered more ‘autochthonous’ than any other in respect of its presence in the subcontinent.”
We finally jump to 2006 and end with two studies. The first was headed by Indian biologist Sanghamitra Sengupta and involved fourteen other co-authors, including L. Cavalli-Sforza, Partha P. Majumder, and P. A. Underhill.17 Based on 728 samples covering 36 Indian populations, it announced in its very title how its findings revealed a “Minor Genetic Influence of Central Asian Pastoralists,” i.e. of the mythical Indo- Aryans, and stated its general agreement with the previous study. For instance, the authors rejected the identification of some Y-DNA genetic markers with an “Indo- European expansion,” an identification they called “convenient but incorrect ... overly simplistic.” To them, the subcontinent’s genetic landscape was formed much earlier than the dates proposed for an Indo-Aryan immigration: “The influence of Central Asia on the pre-existing gene pool was minor. ... There is no evidence whatsoever to conclude that Central Asia has been necessarily the recent donor and not the receptor of the R1a lineages.” This is also highly suggestive (the R1a lineages being a different way to denote the haplogroup M17).
Finally, and significantly, this study indirectly rejected a “Dravidian” authorship of the Indus-Sarasvati civilization, since it noted, “Our data are also more consistent with a peninsular origin of Dravidian speakers than a source with proximity to the Indus....” They found, in conclusion, “overwhelming support for an Indian origin of Dravidian speakers.”
Another Indian biologist, Sanghamitra Sahoo, headed eleven colleagues, including T. Kivisild and V. K. Kashyap, for a study of the Y-DNA of 936 samples covering 77 Indian populations, 32 of them tribes.18 The authors left no room for doubt:
“The sharing of some Y-chromosomal haplogroups between Indian and Central Asian populations is most parsimoniously explained by a deep, common ancestry between the two regions, with diffusion of some Indian- specific lineages northward.”
So the southward gene flow that had been imprinted on our minds for two centuries was wrong, after all: the flow was out of, not into, India. The authors continue:
“The Y-chromosomal data consistently suggest a largely South Asian origin for Indian caste communities and therefore argue against any major influx, from regions north and west of India, of people associated either with the development of agriculture or the spread of the Indo-Aryan language family.”
The last of the two rejected associations is that of the Indo-Aryan expansion; the first, that of the spread of agriculture, is the well-known thesis of Colin Renfrew,19 which traces Indo-European origins to the beginnings of agriculture in Anatolia, and sees Indo-Europeans entering India around 9000 BP, along with agriculture: Sanghamitra Sahoo et al. see no evidence of this in the genetic record.
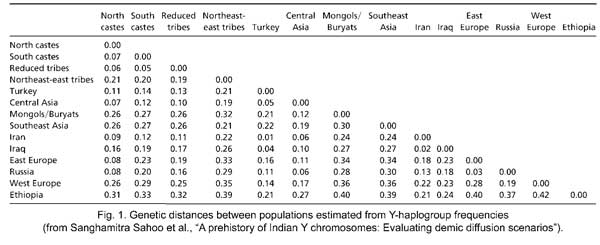
The same data allow the authors to construct an eloquent table of genetic distances between several populations, based on Y-haplogroups (Fig. 2). We learn from it, for instance, that “the caste populations of ‘north’ and ‘south’ India are not particularly more closely related to each other (average Fst value = 0.07) than they are to the tribal groups (average Fst value = 0.06),” an important confirmation of earlier studies. In particular, “Southern castes and tribals are very similar to each other in their Y-chromosomal haplogroup compositions.” As a result, “it was not possible to confirm any of the purported differentiations between the caste and tribal pools,” a momentous conclusion that directly clashes with the Aryan paradigm, which imagined Indian tribes as adivasis and the caste Hindus as descendants of Indo-Aryans invaders or immigrants.
In reality, we have no way, today, to determine who in India is an “adi”-vasi, but enough data to reject this label as misleading and unnecessarily divisive.
![genetic-distance]()
Conclusions
It is, of course, still possible to find genetic studies trying to interpret differences between North and South Indians or higher and lower castes within the invasionist framework, but that is simply because they take it for granted in the first place. None of the nine major studies quoted above lends any support to it, and none proposes to define a demarcation line between tribe and caste. The overall picture emerging from these studies is, first, an unequivocal rejection of a 3500-BP arrival of a “Caucasoid” or Central Asian gene pool. Just as the imaginary Aryan invasion / migration left no trace in Indian literature, in the archaeological and the anthropological record, it is invisible at the genetic level. The agreement between these different fields is remarkable by any standard, and offers hope for a grand synthesis in the near future, which will also integrate agriculture and linguistics.
Secondly, they account for India’s considerable genetic diversity by using a time- scale not of a few millennia, but of 40,000 or 50,000 years. In fact, several experts, such as Lluís Quintana-Murci,20 Vincent Macaulay,21 Stephen Oppenheimer,22 Michael Petraglia,23 and their associates, have in the last few years proposed that when Homo sapiens migrated out of Africa, he first reached South-West Asia around 75,000 BP, and from here, went on to other parts of the world. In simple terms, except for Africans, all humans have ancestors in the North-West of the Indian peninsula. In particular, one migration started around 50,000 BP towards the Middle East and Western Europe:
“indeed, nearly all Europeans — and by extension, many Americans — can trace their ancestors to only four mtDNA lines, which appeared between 10,000 and 50,000 years ago and originated from South Asia.” 24
Oppenheimer, a leading advocate of this scenario, summarizes it in these words:
“For me and for Toomas Kivisild, South Asia is logically the ultimate origin of M17 and his ancestors; and sure enough we find the highest rates and greatest diversity of the M17 line in Pakistan, India, and eastern Iran, and low rates in the Caucasus. M17 is not only more diverse in South Asia than in Central Asia, but diversity characterizes its presence in isolated tribal groups in the south, thus undermining any theory of M17 as a marker of a ‘male Aryan invasion’ of India. One average estimate for the origin of this line in India is as much as 51,000 years. All this suggests that M17 could have found his way initially from India or Pakistan, through Kashmir, then via Central Asia and Russia, before finally coming into Europe.”25
We will not call it, of course, an “Indian invasion” of Europe; in simple terms, India acted “as an incubator of early genetic differentiation of modern humans moving out of Africa.”26
Genetics is a fast-evolving discipline, and the studies quoted above are certainly not the last word; but they have laid the basis for a wholly different perspective of Indian populations, and it is most unlikely that we will have to abandon it to return to the crude racial nineteenth-century fallacies of Aryan invaders and Dravidian autochthons. Neither have any reality in genetic terms, just as they have no reality in archaeological or cultural terms. In this sense, genetics is joining other disciplines in helping to clean the cobwebs of colonial historiography. If some have a vested interest in patching together the said cobwebs so they may keep cluttering our history textbooks, they are only delaying the inevitable.
*
References & Notes
1
Franz Boas, Race, Language and Culture (New York: Macmillan, 1912).
2
Ashley Montagu, Man’s most dangerous myth: The fallacy of race (New York: Columbia
University Press, 1942).
3
Let us mention three important papers: (1) B. E. Hemphill, J. R. Lukacs & K. A. R. Kennedy,
“Biological adaptations and affinities of the Bronze Age Harappans,” in Harappa
Excavations 1986-1990: A Multidisciplinary Approach to Third Millennium Urbanism, ed.
R. H. Meadow (Madison: Prehistory Press, 1991), pp. 137-182. (2) Kenneth A. R. Kennedy,
“Have Aryans been identified in the prehistoric skeletal record from South Asia?” in The
Indo-Aryans of Ancient South Asia, ed. George Erdosy (Berlin & New York: Walter de
Gruyter, 1995), pp. 32-66. (3) Brian E. Hemphill, Alexander F. Christensen & S. I.
Mustafakulov, “Trade or Travel: An Assessment of Interpopulational Dynamics among
Bronze Age Indo-Iranian Populations,” South Asian Archaeology, 1995, ed. Raymond
Allchin & Bridget Allchin (New Delhi: Oxford & IBH Publishing, 1997), vol. 2, pp. 855-
871.
4
See for instance Michael Witzel, “Autochthonous Aryans? The Evidence from Old Indian
and Iranian Texts,” Electronic Journal of Vedic Studies, vol. 7 (2001), No. 3 (25 May), § 8.
5
For a fuller discussion of this and other paradoxes of the Aryan invasion theory, see Michel
Danino, L’Inde et l’invasion de nulle part: le dernier repaire du mythe aryen (Paris: Les
Belles Lettres, 2006), forthcoming in English as The Invasion That Never Was, 3rd ed.
6
Ram Sharan Sharma, Advent of the Aryans in India (New Delhi: Manohar, 2001), p. 52.
7
See a few examples in The Indian Human Heritage, ed. D. Balasubramanian & N. Appaji
Genetics and the Aryan Debate / p. 12
Rao (Hyderabad: Universities Press, 1998).
8
This is the case of L. L. Cavalli-Sforza, of Stanford University, co-author of a “classic” work
which, as regards India, did not dare to question the invasionist framework: L. L. Cavalli-
Sforza, P. Menozzi & A. Piazza, The History and Geography of Human Genes (Princeton:
Princeton University Press, 1994); twelve years later, Cavalli-Sforza co-authored two papers
that rejected this framework, see notes 15 & 17 below. Another case is that of the Indian
biologist Partha P. Majumder (see notes 12 & 17 below).
9
T. Kivisild, M. J. Bamshad, K. Kaldma, M. Metspalu, E. Metspalu, M. Reidla, S. Laos, J.
Parik, W. S. Watkins, M. E. Dixon, S. S. Papiha, S. S. Mastana, M. R. Mir, V. Ferak, R.
Villems, “Deep common ancestry of Indian and western-Eurasian mitochondrial DNA
lineages” in Current Biology, 18 November 1999, 9(22):1331-4. (Most of the articles quoted
in this paper are available on the Internet; to locate them, enter their full title in a good search
engine.)
10
“Caucasoid” is a nineteenth-century term for a member of the white race, coined at a time
when the Caucasus was thought to be the homeland of the Indo-Europeans. The term has no
scientific meaning but has stuck, and is still used occasionally by biologists, although, as
further quotations will show, often within quotation marks, as a reminder of its inadequacy.
11
T. R. Disotell, “Human evolution: the southern route to Asia” in Current Biology, vol. 9, No.
24, 16 December 1999, pp. R925-928(4).
12
Susanta Roychoudhury, Sangita Roy, Badal Dey, Madan Chakraborty, Monami Roy, Bidyut
Roy, A. Ramesh, N. Prabhakaran, M. V. Usha Rani, H. Vishwanathan, Mitashree Mitra,
Samir K. Sil & Partha P. Majumder, “Fundamental genomic unity of ethnic India is revealed
by analysis of mitochondrial DNA,” Current Science, vol. 79, No. 9, 10 November 2000, pp.
1182-1192.
13
Toomas Kivisild, Surinder S. Papiha, Siiri Rootsi, Jüri Parik, Katrin Kaldma, Maere Reidla,
Sirle Laos, Mait Metspalu, Gerli Pielberg, Maarja Adojaan, Ene Metspalu, Sarabjit S.
Mastana, Yiming Wang, Mukaddes Golge, Halil Demirtas, Eckart Schnakenberg, Gian
Franco de Stefano, Tarekegn Geberhiwot, Mireille Claustres & Richard Villems, “An Indian
Ancestry: a Key for Understanding Human Diversity in Europe and Beyond”, ch. 31 of
Archaeogenetics: DNA and the population prehistory of Europe, ed. Colin Renfrew & Katie
Boyle (Cambridge: McDonald Institute for Archaeological Research, 2000), pp. 267-275.
14
Toomas Kivisild, Siiri Rootsi, Mait Metspalu, Ene Metspalu, Juri Parik, Katrin Kaldma,
Esien Usanga, Sarabjit Mastana, Surinder S. Papiha & Richard Villems, “The Genetics of
Language and Farming Spread in India,” ch. 17 in Examining the farming/language
dispersal hypothesis, eds. Peter Bellwood & Colin Renfrew (Cambridge: McDonald Institute
for Archaeological Research, 2003), pp. 215–222. Italics in one of the quotations are in the
original.
15
T. Kivisild, S. Rootsi, M. Metspalu, S. Mastana, K. Kaldma, J. Parik, E. Metspalu, M.
Adojaan, H.-V. Tolk, V. Stepanov, M. Gölge, E. Usanga, S. S. Papiha, C. Cinnioglu, R. King,
L. Cavalli-Sforza, P. A. Underhill & R. Villems, “The Genetic Heritage of the Earliest
Settlers Persists Both in Indian Tribal and Caste Populations,” American Journal of Human
Genetics and the Aryan Debate / p. 13
Genetics 72(2):313-32, 2003.
16
Mait Metspalu, Toomas Kivisild, Ene Metspalu, Jüri Parik, Georgi Hudjashov, Katrin
Kaldma, Piia Serk, Monika Karmin, Doron M Behar, M Thomas P Gilbert, Phillip Endicott,
Sarabjit Mastana, Surinder S. Papiha, Karl Skorecki, Antonio Torroni & Richard Villem,
“Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped
during the initial settlement of Eurasia by anatomically modern humans,” BMC Genetics
2004, 5:26.
17
Sanghamitra Sengupta, Lev A. Zhivotovsky, Roy King, S. Q. Mehdi, Christopher A.
Edmonds, Cheryl-Emiliane T. Chow, Alice A. Lin, Mitashree Mitra, Samir K. Sil, A.
Ramesh, M. V. Usha Rani, Chitra M. Thakur, L. Luca Cavalli-Sforza, Partha P. Majumder, &
Peter A. Underhill, “Polarity and Temporality of High-Resolution Y-Chromosome
Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal
Minor Genetic Influence of Central Asian Pastoralists,” American Journal of Human
Genetics, February 2006; 78(2):202-21. (Italics in one of the quotations are mine.)
18
Sanghamitra Sahoo, Anamika Singh, G. Himabindu, Jheelam Banerjee, T. Sitalaximi, Sonali
Gaikwad, R. Trivedi, Phillip Endicott, Toomas Kivisild, Mait Metspalu, Richard Villems, &
V. K. Kashyap, “A prehistory of Indian Y chromosomes: Evaluating demic diffusion
scenarios,” Proceedings of the National Academy of Sciences, 24 January 2006, vol. 103,
No. 4, pp. 843–848. (Italics in one of the quotations are mine.)
19
Colin Renfrew, Archaeology and Language: the Puzzle of Indo-European Origins (London:
Penguin Books, 1989).
20
Lluís Quintana-Murci, Raphaëlle Chaix, R. Spencer Wells, Doron M. Behar, Hamid Sayar,
Rosaria Scozzari, Chiara Rengo, Nadia Al-Zahery, Ornella Semino, A. Silvana Santachiara-
Benerecetti, Alfredo Coppa, Qasim Ayub, Aisha Mohyuddin, Chris Tyler-Smith, S. Qasim
Mehdi, Antonio Torroni, & Ken McElreavey, “Where West Meets East: The Complex
mtDNA Landscape of the Southwest and Central Asian Corridor,” American Journal of
Human Genetics 74(5):827-45, May 2004.
21
Vincent Macaulay, Catherine Hill, Alessandro Achilli, Chiara Rengo, Douglas Clarke,
William Meehan, James Blackburn, Ornella Semino, Rosaria Scozzari, Fulvio Cruciani, Adi
Taha, Norazila Kassim Shaari,6 Joseph Maripa Raja, Patimah Ismail, Zafarina Zainuddin,
William Goodwin, David Bulbeck, Hans-Jürgen Bandelt, Stephen Oppenheimer, Antonio
Torroni, Martin Richards, “Single, Rapid Coastal Settlement of Asia Revealed by Analysis of
Complete Mitochondrial Genomes,” Science 13 May 2005, vol. 308, No. 5724, pp. 1034-36.
22
Stephen Oppenheimer, The Real Eve: Modern Man’s Journey out of Africa (New York:
Carroll & Graf Publishers, 2003). See an introduction to Oppenheimer’s theory on the
website:
www.bradshawfoundation.com.
23
Hannah V. A. James & Michael D. Petraglia, “Modern Human Origins and the Evolution of
Behavior in the Later Pleistocene Record of South Asia,” Current Anthropology vol. 46,
Supplement, December 2005, pp. S3-S27.
Genetics and the Aryan Debate / p. 14
24
William F. Allman, “Eve Explained: How Ancient Humans Spread Across the Earth” (on the
website of Discovery Channel, 21 August 2004).
25
Stephen Oppenheimer, The Real Eve, op. cit., p. 152.
26
![siva temples on straight line]()











.jpg?rect=0,0,600,338&w=661&q=60&fmt=pjpeg&auto=format)
.jpg?w=661&q=60&fmt=pjpeg&auto=format)
















 Claudia Waedlich hails from Germany. She is a literary writer, lawyer, politician and part of The Pashtun Times International News Network. She can be reached at
Claudia Waedlich hails from Germany. She is a literary writer, lawyer, politician and part of The Pashtun Times International News Network. She can be reached at

 Despite his
Despite his  And whenever there is compromise, there is bound to be pitfalls.
And whenever there is compromise, there is bound to be pitfalls.

Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply
Reply